 |
Case Study from the Cambridge Structural Database |
 |
Case Study from the Cambridge Structural Database |
Case Study from the Cambridge Structural Database
In this example we search the CDS for structures with the following three restrictions:
Results
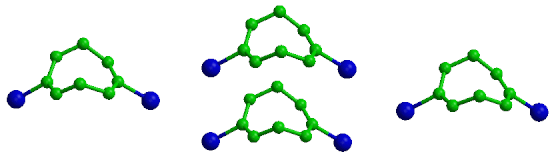
The search produces 6 structures that fulfill the requirements of this search.
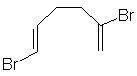
The actual type of information produced for one of these structures is illustrated for C8H10Br2. The red italicised text is for explanation purposes only and is not given in the actual output.
1D Information
TERSOY Reference Code
1,5-Dibromocyclo-octa-1,5-diene Chemical Name
C8 H10 Br2 Molecular Formula
A.Davila,M.L.McLaughlin,F.R.Fronczek,S.F.Watkins Authors
Acta Crystallogr.,Sect.C(Cr.Str.Comm.), 53, 84,1997 Journal Reference
2D Information

3D information
3D Molecular Structure
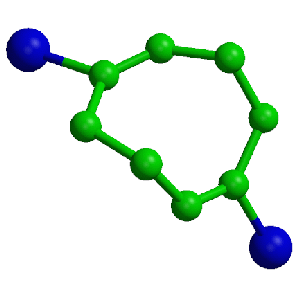
The data from which the 3D structures are derived may be output as text files
with the following format:
(a)TERSOY 23970221 13 9 20 0 0 4 3 10 10 0 22132100000010000000000097
(b)188917 (c)46858 (d)12
(i)R=0.0380 (j)CIF entry bk1224.
211 0121 0112 0011 0121 0110 6211 6121 6112 0011 6121 6110 6
C 68H 23BR121
(k)BR1 66801 48532 73984 C1 58660 24130 74170 C2 50860 18670 62730
C3 43440 -170 59990 C4 62750 14480 88000 H1 49800 28900 55100
H2 39800 -5000 50700 H3 45800 -19100 65000 H4 65200 32100 93100
H5 67900 1700 90800 (l)C4A 37250 14480 62000 C1A 41340 24130 75830
BR1A 33199 48532 76016 C2A 49140 18670 87270 C3A 56560 -170 90010
H2A 60200 -5000 99300 H3A 54200 -19100 85000 H1A 50200 28900 94900
H4A 34800 32100 56900 H5A 32100 1700 59200
2 3 4 7 2 3 0 4 5 5 4111212 515151411111415
where (referring to the red letters in the above table):
Geometric parameters from the above coordinates can be derived, such as, bond
lengths and bond angles, and are output in the following format:
TERSOY geometric data.
Bond lengths
Br1 C1 1.928 C1 C2 1.314 C1 C4 1.500 C2 C3 1.504
C2 H1 0.991 C3 H2 0.965 C3 H3 1.022 C3 C4A 1.508
C4 H4 0.975 C4 H5 1.002 C4 C3A 1.508 C4A C1A 1.500
C4A H4A 0.975 C4A H5A 1.002 C1A Br1A 1.928 C1A C2A 1.314
C2A H1A 0.991 C2A C3A 1.504 C3A H2A 0.965 C3A H3A 1.022
Bond angles
Br1 C1 C2 117.3 Br1 C1 C4 110.9 C2 C1 C4 131.8
C1 C2 C3 128.3 C1 C2 H1 114.3 C3 C2 H1 117.4
C2 C3 H2 106.3 C2 C3 H3 112.2 C2 C3 C4A 113.7
H2 C3 H3 105.9 H2 C3 C4A 106.1 H3 C3 C4A 111.9
C1 C4 H4 102.8 C1 C4 H5 106.5 C1 C4 C3A 116.6
H4 C4 H5 108.0 H4 C4 C3A 111.7 H5 C4 C3A 110.6
C3 C4A C1A 116.6 C3 C4A H4A 111.7 C3 C4A H5A 110.6
C1A C4A H4A 102.8 C1A C4A H5A 106.5 H4A C4A H5A 108.0
C4A C1A Br1A 110.9 C4A C1A C2A 131.8 Br1A C1A C2A 117.3
C1A C2A H1A 114.3 C1A C2A C3A 128.3 H1A C2A C3A 117.4
C4 C3A H2A 106.1 C4 C3A H3A 111.9 C4 C3A C2A 113.7
H2A C3A H3A 105.9 H2A C3A C2A 106.3 H3A C3A C2A 112.2
Statistical Analysis
Below is an example of the type of simple statistical analysis that is
possible using the CDS.
The analysis is of the C-Br bond lengths in the 6 structures containing the
fragment we discussed above.
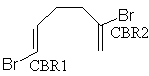
Nfrag Refcode CBR1 CBR2 CBR
1 AERTHNa 1.918 1.889 1.904
2 AERTHNa 1.838 1.904 1.871
3 ARPLYS 1.894 1.879 1.886
4 EXBSUN 1.910 1.911 1.911
5 GAJVOCb 1.846 1.848 1.847
6 GAJVOCb 1.895 1.848 1.871
7 JAWVOS 1.903 1.903 1.903
8 TERSOY 1.928 1.928 1.928
Nent 6
Nobs 8 8 8
Mean 1.891 1.889 1.890
SDSample 0.033 0.029 0.026
SDMean 0.012 0.010 0.009
Minimum 1.838 1.848 1.847
Maximum 1.928 1.928 1.928
aThere are 2 entries for AERTHN as there are 2 crystallographically
different fragments in the same molecule.
Histogram for parameter CBR1
(Bond length range) (Number of bonds in range)
1.835 TO 1.840 1 *
1.840 TO 1.845 0
1.845 TO 1.850 1 *
1.850 TO 1.855 0
1.855 TO 1.860 0
1.860 TO 1.865 0
1.865 TO 1.870 0
1.870 TO 1.875 0
1.875 TO 1.880 0
1.880 TO 1.885 0
1.885 TO 1.890 0
1.890 TO 1.895 2 **
1.895 TO 1.900 0
1.900 TO 1.905 1 *
1.905 TO 1.910 1 *
1.910 TO 1.915 0
1.915 TO 1.920 1 *
1.920 TO 1.925 0
1.925 TO 1.930 1 *
Histogram for parameter CBR2
1.846 TO 1.850 2 **
1.850 TO 1.854 0
1.854 TO 1.858 0
1.858 TO 1.862 0
1.862 TO 1.866 0
1.866 TO 1.870 0
1.870 TO 1.874 0
1.874 TO 1.878 0
1.878 TO 1.882 1 *
1.882 TO 1.886 0
1.886 TO 1.890 1 *
1.890 TO 1.894 0
1.894 TO 1.898 0
1.898 TO 1.902 0
1.902 TO 1.906 2 **
1.906 TO 1.910 0
1.910 TO 1.914 1 *
1.914 TO 1.918 0
1.918 TO 1.922 0
1.922 TO 1.926 0
1.926 TO 1.930 1 *
Histogram for parameter CBR
1.846 TO 1.850 1 *
1.850 TO 1.854 0
1.854 TO 1.858 0
1.858 TO 1.862 0
1.862 TO 1.866 0
1.866 TO 1.870 0
1.870 TO 1.874 2 **
1.874 TO 1.878 0
1.878 TO 1.882 0
1.882 TO 1.886 0
1.886 TO 1.890 1 *
1.890 TO 1.894 0
1.894 TO 1.898 0
1.898 TO 1.902 0
1.902 TO 1.906 2 **
1.906 TO 1.910 0
1.910 TO 1.914 1 *
1.914 TO 1.918 0
1.918 TO 1.922 0
1.922 TO 1.926 0
1.926 TO 1.930 1 *
| © Copyright 1997-2006. Birkbeck College, University of London. | Author(s): Martin Attfield |